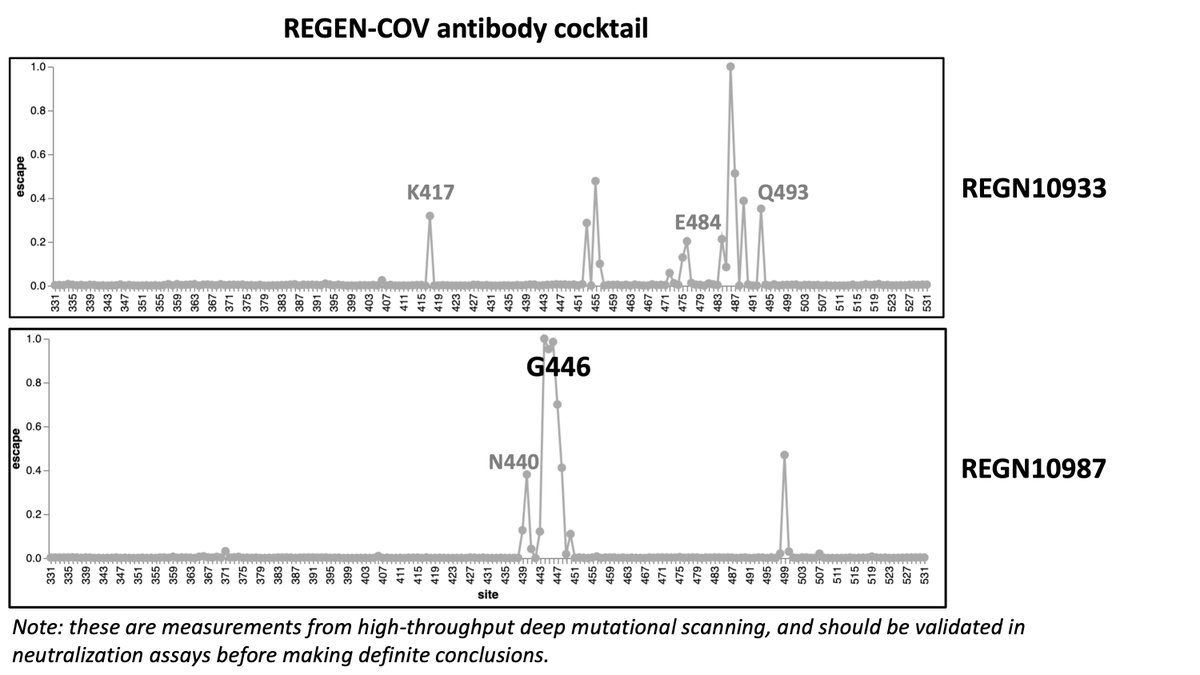
Here's how mutations in #SARSCoV2 Nu variant (B.1.1.529) will affect polyclonal and monoclonal antibodies targeting RBD. These assessments based on deep-mutational scanning experiments; underlying data can be explored interactively at jbloomlab.github.io (1/n)
Another way to assess polyclonal escape is how many epitope classes affected (nature.com). We do this using epitope scheme of @bjorkmanlab @cobarnes27 as adopted by @AllieGreaney. In this scheme, three potently neutralizing epitopes: class 1, 2, class 3. (3/n)
Importantly, this does *not* mean Nu variant will fully escape vaccine- or infection-elicited antibodies. @PaulBieniasz @theodora_nyc have shown takes many many mutations to fully escape neutralization (nature.com), & there are also T-cells, non-neut Abs, etc. (5/n)
But I'd expect the Nu variant to cause more of a hit on vaccine- and infection-elicited antibody neutralization than anything we've seen so far. (6/n)
You can explore other antibodies that might be of interest to you at jbloomlab.github.io. Importantly, all above results from high-throughput deep mutational scanning, and need to be validated in traditional experiments for high confidence. (10/n)
Also note large antigenic change does not mean Nu will necessarily spread & outcompete other variants. That will also depend on its transmissibility, which has been discussed by @Tuliodna & others (eg, and for which data remains preliminary. (11/n)
As @trvrb discussed in excellent recent thread, selection on variants so far may be dominated more by transmissibility than antigenic selection ( But I'm not as sanguine that antigenic selection isn't also playing substantial role... (12/n)
Reason I say that is comparison of Nu variant to BANAL-20-52, a SARS-related CoV isolated from bats. If we compare both BANAL-20-52 and Nu to Wuhan-Hu-1, Nu has *many* more mutations that strongly affect antigenicity ( (13/n)
If selection was mostly for transmissibility, I'd expect sites of divergence of Nu and BANAL-20-52 relative to Wuhan-Hu-1 to perhaps be similarly distributed with respect to antigenic sites. But instead, Nu mutations much more focused in major antigenic sites. (14/n)
We can also use deep mutational scanning to assess how mutations in Nu affect ACE2 affinity ( But I suspect works less well than for antigenic mutations discussed above as there's lot more mutational epistasis for ACE2 affinity (eg, N501 & Q498). (15/n)
Important caveat: all of above is based on deep mutational scanning experiments. I'm sure more Nu-specific data will emerge in coming weeks to months. But I think it's useful to use prospective data we already have to calibrate what to expect. (16/n)
Thanks to @AllieGreaney @tylernstarr for doing deep mutational scanning on which above is predicated, & @NussenzweigL @VUMC_Vaccines @seth_zost @Vir_Biotech for sharing the antibodies. And of course the scientists providing rapid information about Nu (
And probably I should have used the variant name B.1.1.529 throughout above thread...
For people interested in G446 mutations, I'm going to link back to some details on this site. Here is an old thread discussing how G446 is a major site of escape in the class 3 epitope where (as of March 2021) mutations were not prevalent:
Also, here are old data showing how for a minority of convalescent individuals, the epitope centered on G446 is immunodominant with respect to serum neutralization:
Also, checkout the awesome CoV-RDB database of Bob Shafer, @Philip_Tzou, @sergeilkp, K Tao which compiles experimental data on G446 (and also whatever other mutation you care about): covdb.stanford.edu
Here are data from one of @AllieGreaney's papers (Fig 5 of science.org) showing that a K417-G446-E484 triple mutant sometimes but not always fully escapes neutralization by RBD-targeted antibodies, and effect is worse for convalescent than mRNA-1273 vaccine sera.
Also adding this as another highly informed view just for balance on question of possible extent of antibody neutralization escape:
جاري تحميل الاقتراحات...